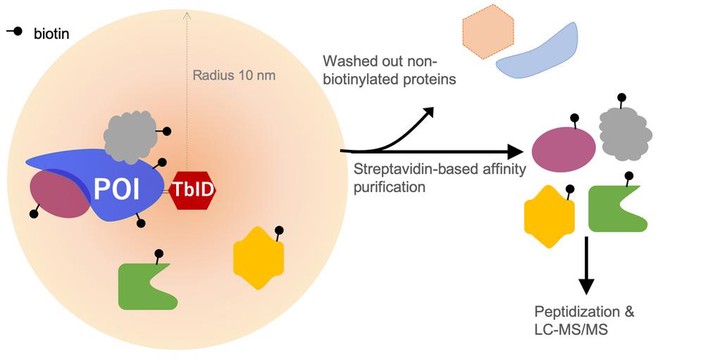
Proximity-Dependent Biotinylation assays coupled to Mass Spectrometry (PDB-MS) has changed the field of Protein–Protein interaction studies. The two enzymes commonly used for Proximity Labeling (PL) come with tradeoffs:
- BioID is slow, requiring tagging times of 18-24 hours
- APEX peroxidase uses substrates that have limited cell permeability and high toxicity
TurboID:
- Greater catalytic efficiency than BioID
- PL in cells in much shorter time windows (10 minutes)
- Non-toxic and easily deliverable Biotin